Text classification: Missing areas¶
[1]:
import warnings
from abc import ABC, abstractmethod
import matplotlib.pyplot as plt
import numpy as np
from numba import NumbaDeprecationWarning, NumbaWarning
from numpy.random import RandomState
from sklearn.exceptions import ConvergenceWarning
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import train_test_split
from sklearn.naive_bayes import MultinomialNB
from sklearn.svm import LinearSVC
from dpemu import runner
from dpemu.dataset_utils import load_newsgroups
from dpemu.filters.text import MissingArea
from dpemu.ml_utils import reduce_dimensions_sparse
from dpemu.nodes.array import Array
from dpemu.plotting_utils import visualize_best_model_params, visualize_scores, visualize_classes, \
print_results_by_model, visualize_confusion_matrices
from dpemu.radius_generators import GaussianRadiusGenerator
warnings.simplefilter("ignore", category=ConvergenceWarning)
warnings.simplefilter("ignore", category=NumbaDeprecationWarning)
warnings.simplefilter("ignore", category=NumbaWarning)
[2]:
def get_data():
data, labels, label_names, dataset_name = load_newsgroups("all", 10)
train_data, test_data, train_labels, test_labels = train_test_split(data, labels, test_size=.2,
random_state=RandomState(42))
return train_data, test_data, train_labels, test_labels, label_names, dataset_name
[3]:
def get_err_root_node():
err_root_node = Array()
err_root_node.addfilter(MissingArea("p", "radius_generator", "missing_value"))
return err_root_node
[4]:
def get_err_params_list():
p_steps = np.linspace(0, .28, num=8)
err_params_list = [{
"p": p,
"radius_generator": GaussianRadiusGenerator(0, 1),
"missing_value": " "
} for p in p_steps]
return err_params_list
[5]:
class Preprocessor:
def __init__(self):
self.random_state = RandomState(0)
def run(self, train_data, test_data, _):
vectorizer = TfidfVectorizer(max_df=0.5, min_df=2, stop_words="english")
vectorized_train_data = vectorizer.fit_transform(train_data)
vectorized_test_data = vectorizer.transform(test_data)
reduced_test_data = reduce_dimensions_sparse(vectorized_test_data, self.random_state)
return vectorized_train_data, vectorized_test_data, {"reduced_test_data": reduced_test_data}
[6]:
class AbstractModel(ABC):
def __init__(self):
self.random_state = RandomState(42)
@abstractmethod
def get_fitted_model(self, train_data, train_labels, params):
pass
def run(self, train_data, test_data, params):
train_labels = params["train_labels"]
test_labels = params["test_labels"]
fitted_model = self.get_fitted_model(train_data, train_labels, params)
predicted_test_labels = fitted_model.predict(test_data)
cm = confusion_matrix(test_labels, predicted_test_labels)
return {
"confusion_matrix": cm,
"predicted_test_labels": predicted_test_labels,
"test_mean_accuracy": round(np.mean(predicted_test_labels == test_labels), 3),
"train_mean_accuracy": fitted_model.score(train_data, train_labels),
}
class MultinomialNBModel(AbstractModel):
def __init__(self):
super().__init__()
def get_fitted_model(self, train_data, train_labels, params):
return MultinomialNB(params["alpha"]).fit(train_data, train_labels)
class LinearSVCModel(AbstractModel):
def __init__(self):
super().__init__()
def get_fitted_model(self, train_data, train_labels, params):
return LinearSVC(C=params["C"], random_state=self.random_state).fit(train_data, train_labels)
[7]:
def get_model_params_dict_list(train_labels, test_labels):
alpha_steps = [10 ** i for i in range(-4, 1)]
C_steps = [10 ** k for k in range(-3, 2)]
model_params_base = {"train_labels": train_labels, "test_labels": test_labels}
return [
{
"model": MultinomialNBModel,
"params_list": [{"alpha": alpha, **model_params_base} for alpha in alpha_steps],
"use_clean_train_data": False
},
{
"model": MultinomialNBModel,
"params_list": [{"alpha": alpha, **model_params_base} for alpha in alpha_steps],
"use_clean_train_data": True
},
{
"model": LinearSVCModel,
"params_list": [{"C": C, **model_params_base} for C in C_steps],
"use_clean_train_data": False
},
{
"model": LinearSVCModel,
"params_list": [{"C": C, **model_params_base} for C in C_steps],
"use_clean_train_data": True
},
]
[8]:
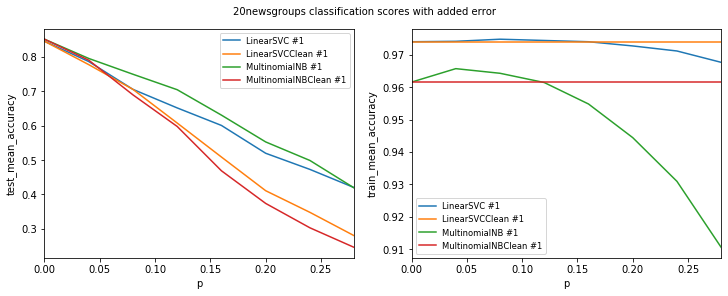
def visualize(df, dataset_name, label_names, test_data):
visualize_scores(
df,
score_names=["test_mean_accuracy", "train_mean_accuracy"],
is_higher_score_better=[True, True],
err_param_name="p",
title=f"{dataset_name} classification scores with added error"
)
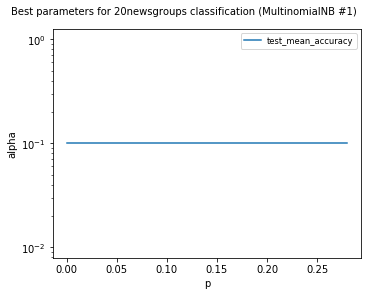
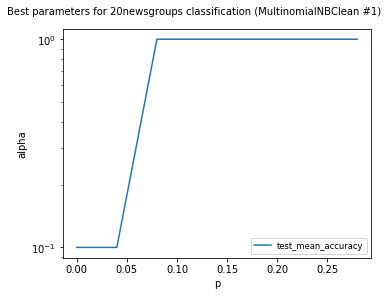
visualize_best_model_params(
df,
"MultinomialNB",
model_params=["alpha"],
score_names=["test_mean_accuracy"],
is_higher_score_better=[True],
err_param_name="p",
title=f"Best parameters for {dataset_name} classification",
y_log=True
)
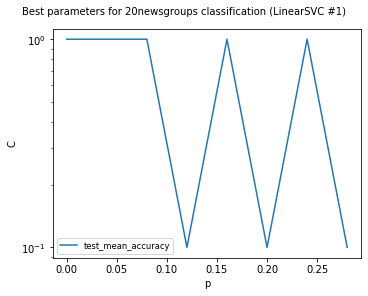
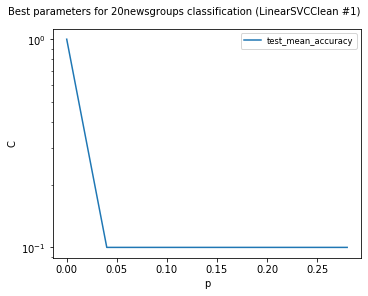
visualize_best_model_params(
df,
"LinearSVC",
model_params=["C"],
score_names=["test_mean_accuracy"],
is_higher_score_better=[True],
err_param_name="p",
title=f"Best parameters for {dataset_name} classification",
y_log=True
)
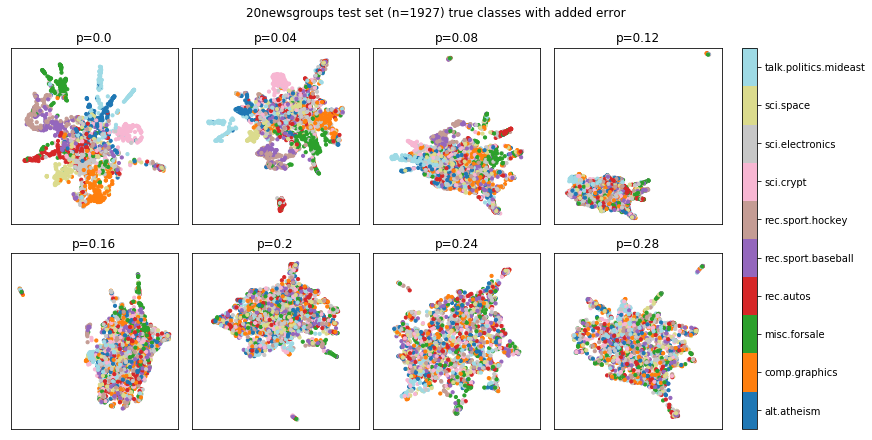
visualize_classes(
df,
label_names,
err_param_name="p",
reduced_data_column="reduced_test_data",
labels_column="test_labels",
cmap="tab20",
title=f"{dataset_name} test set (n={len(test_data)}) true classes with added error"
)
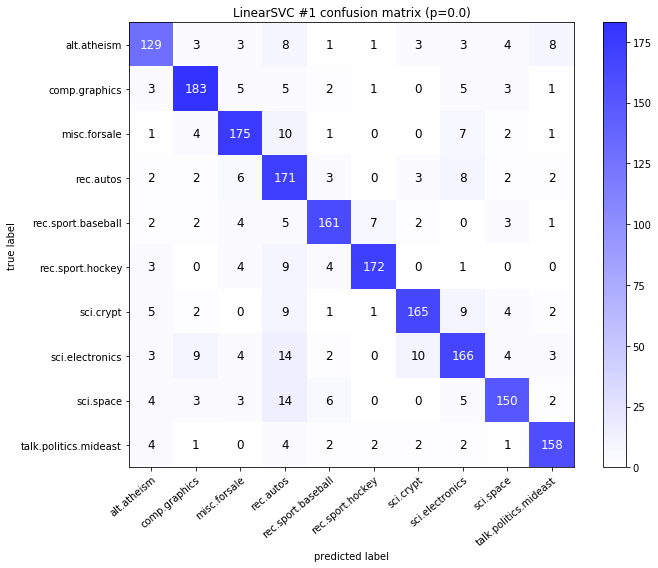
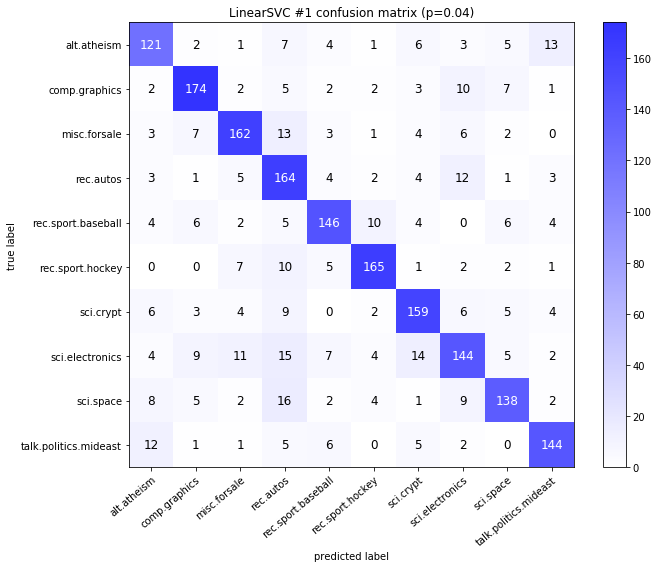
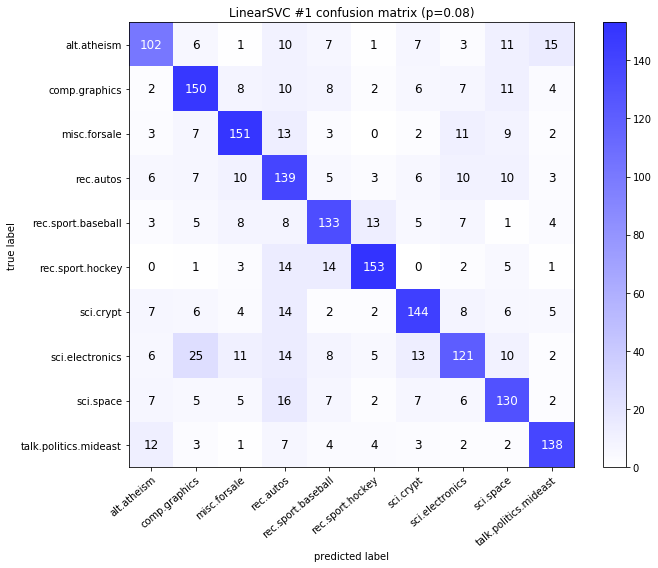
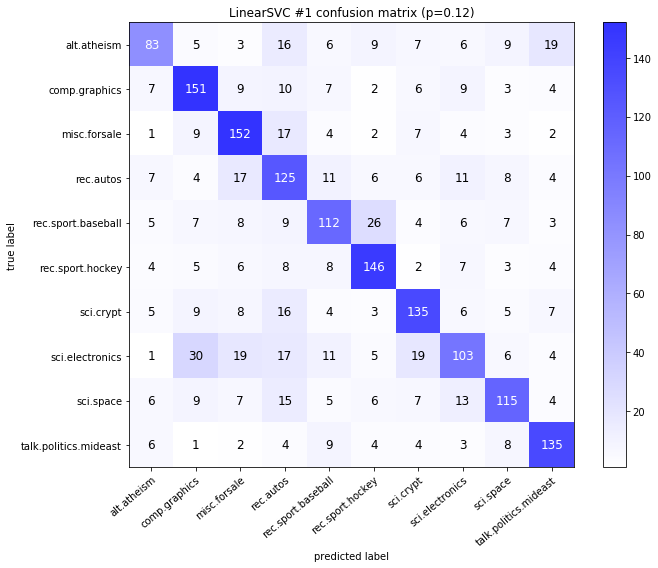
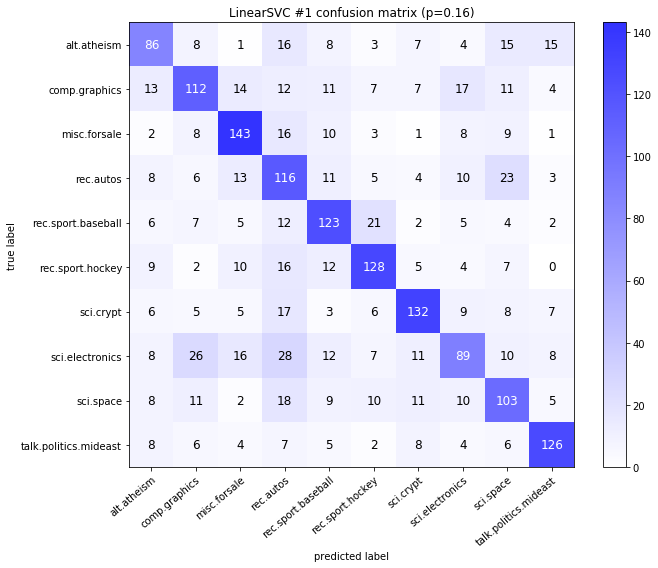
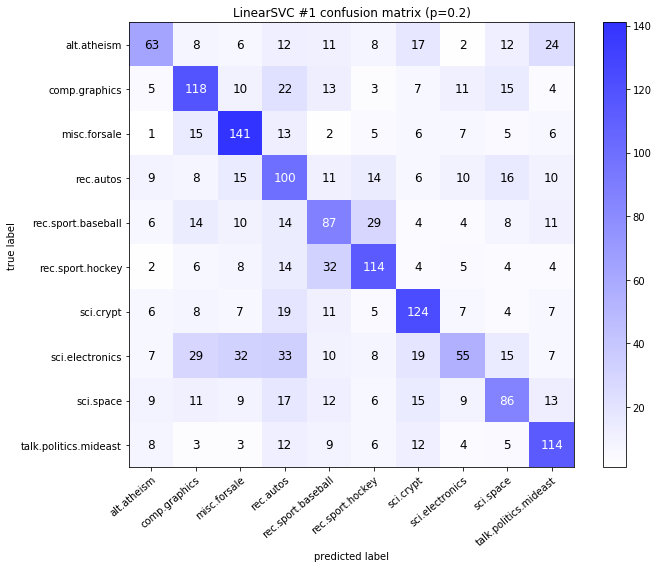
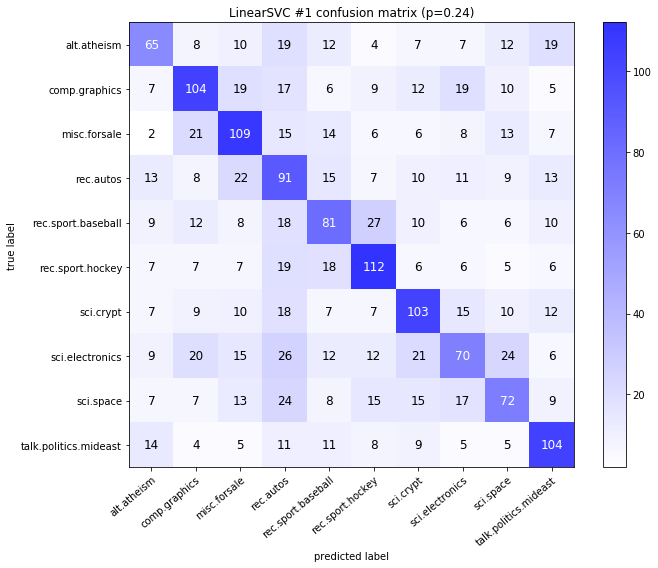
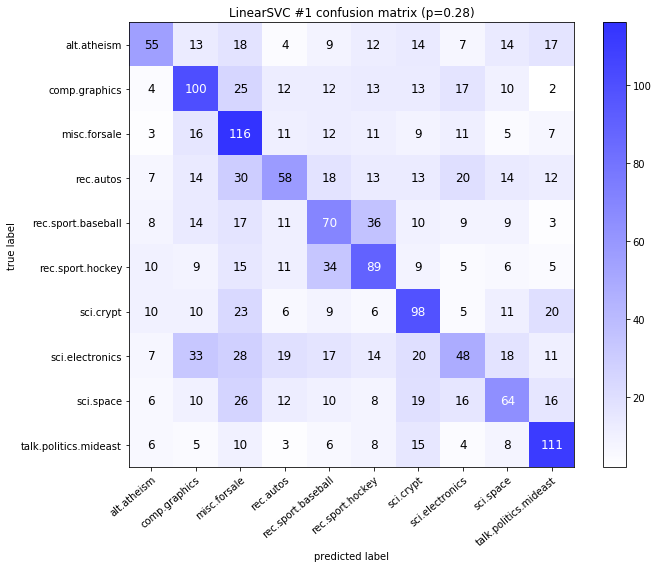
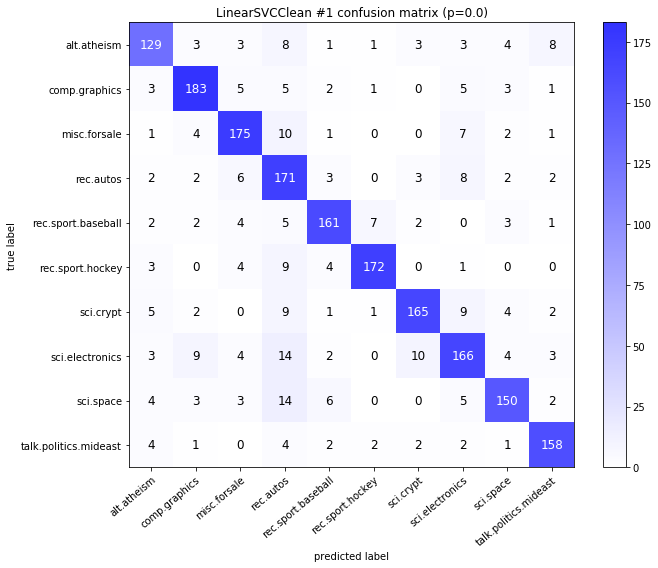
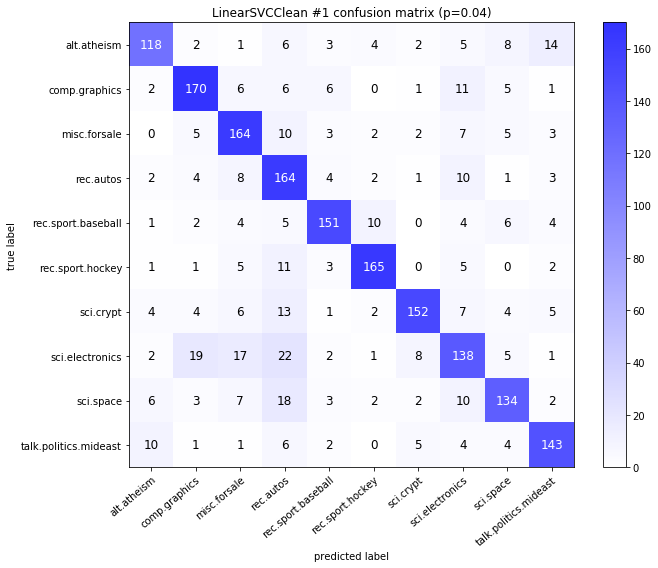
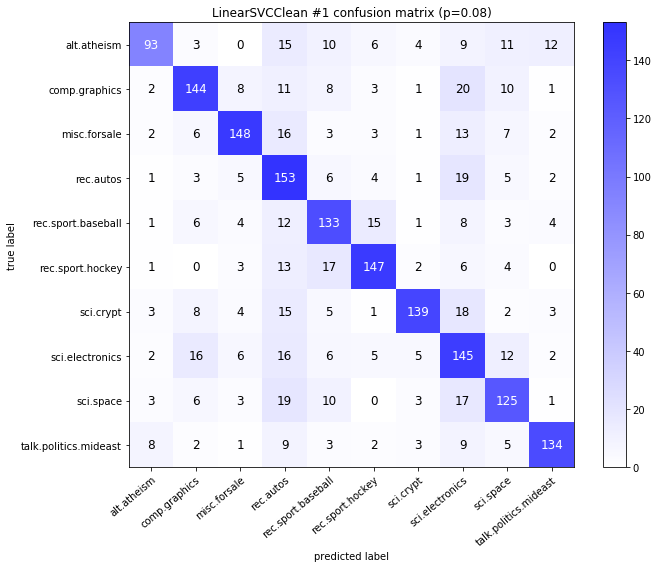
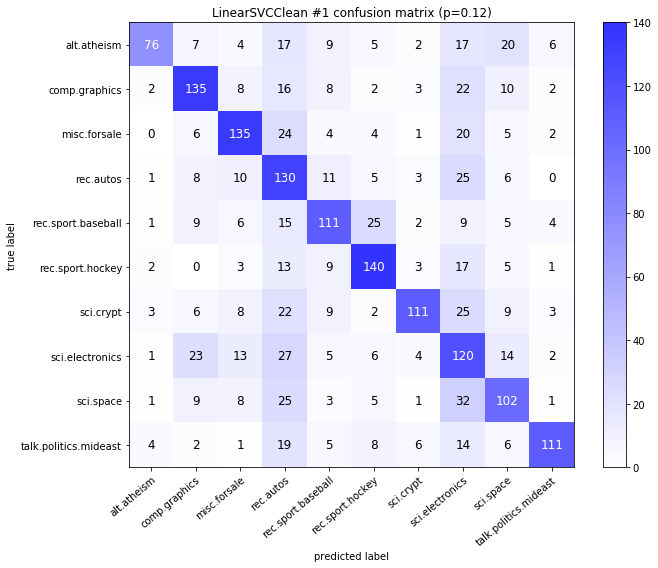
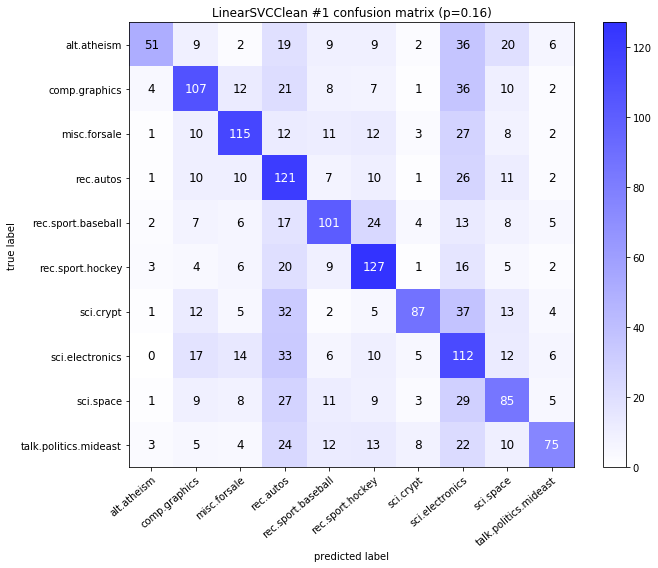
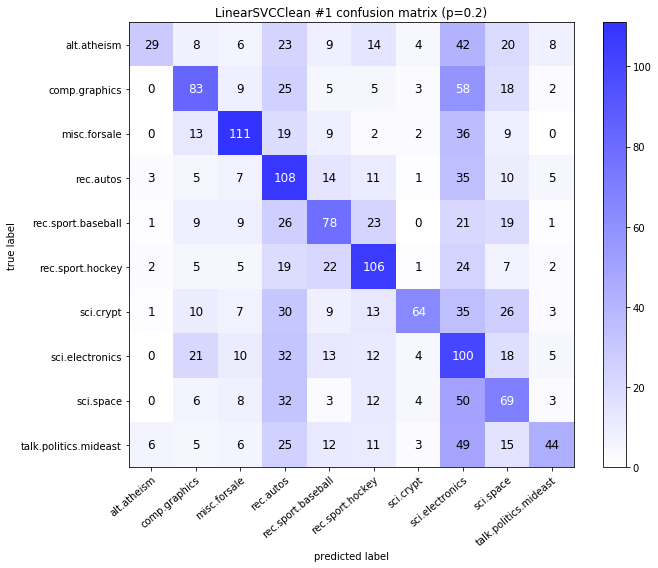
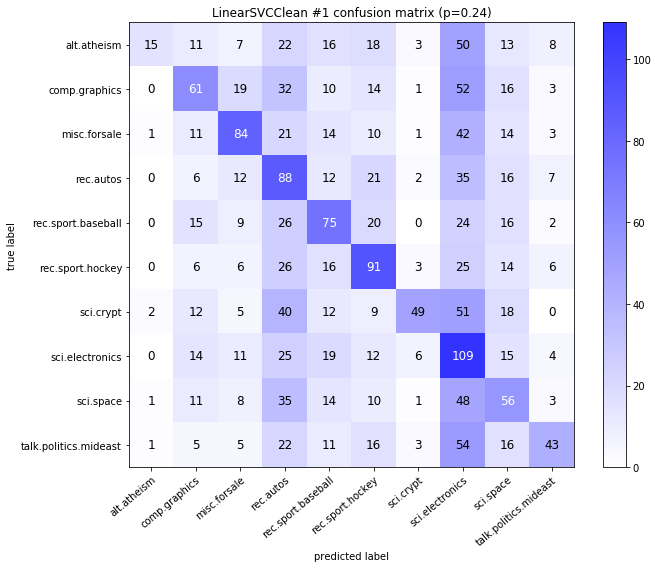
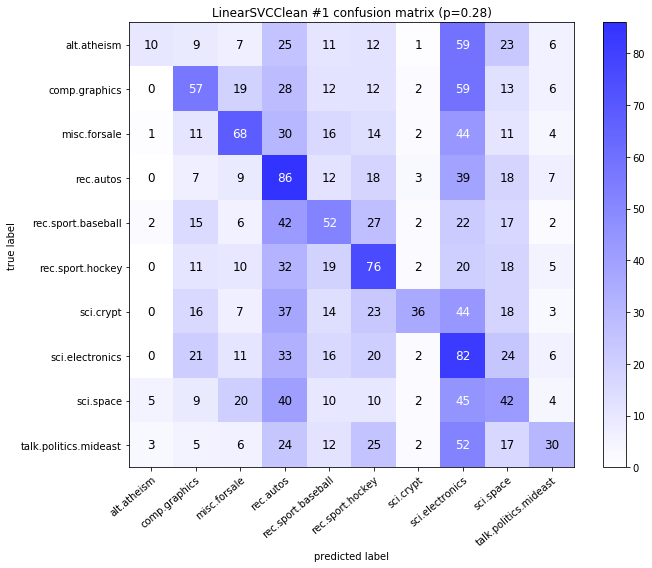
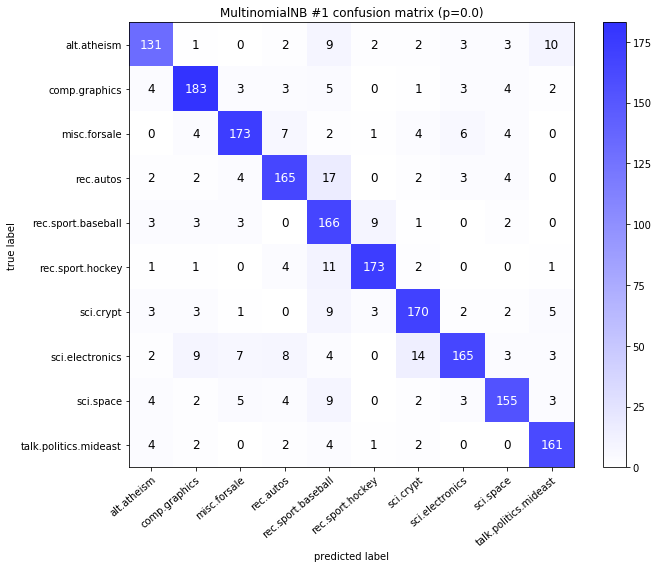
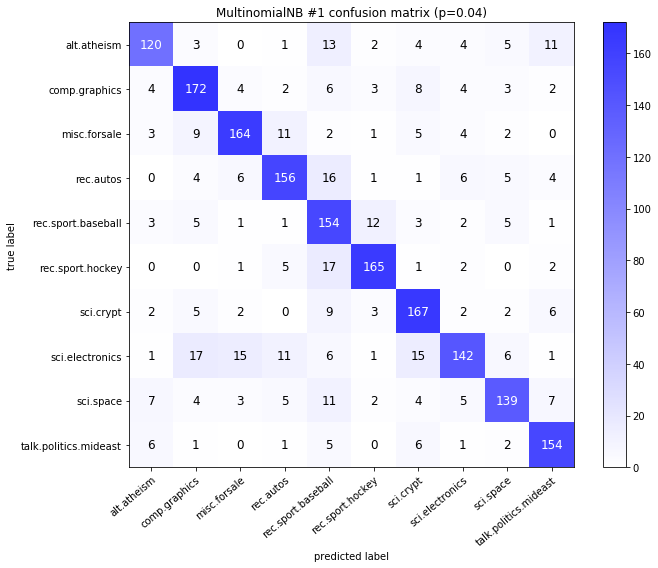
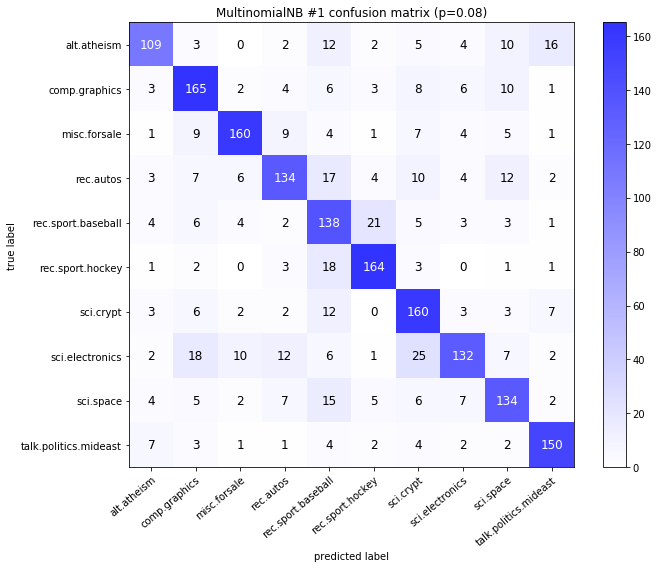
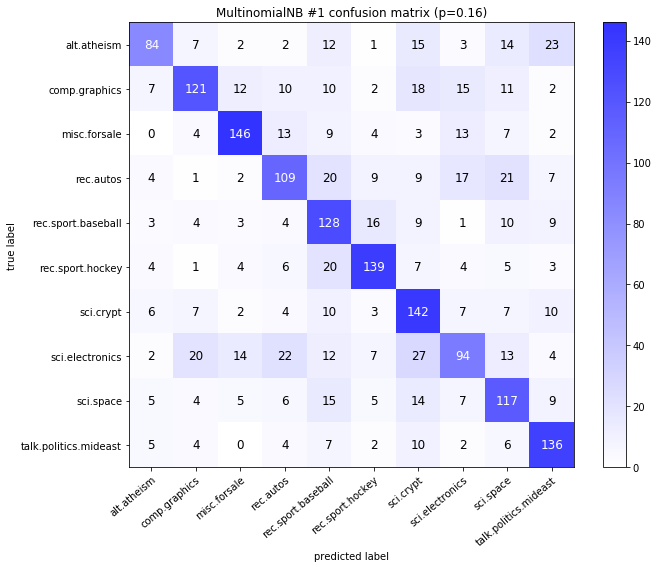
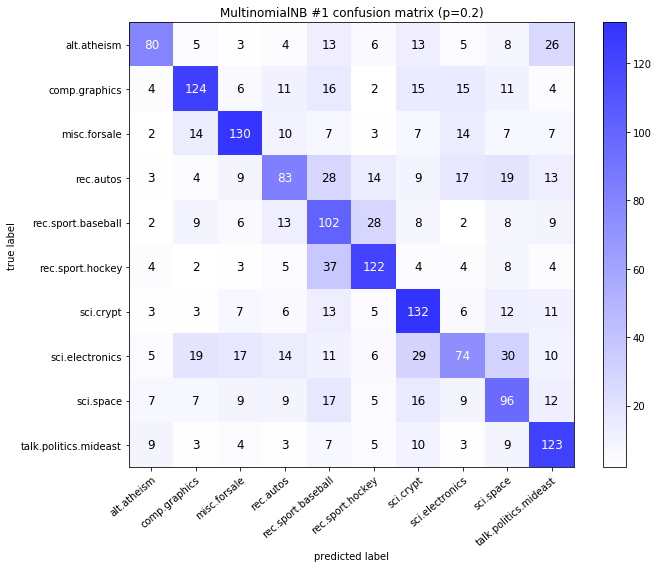
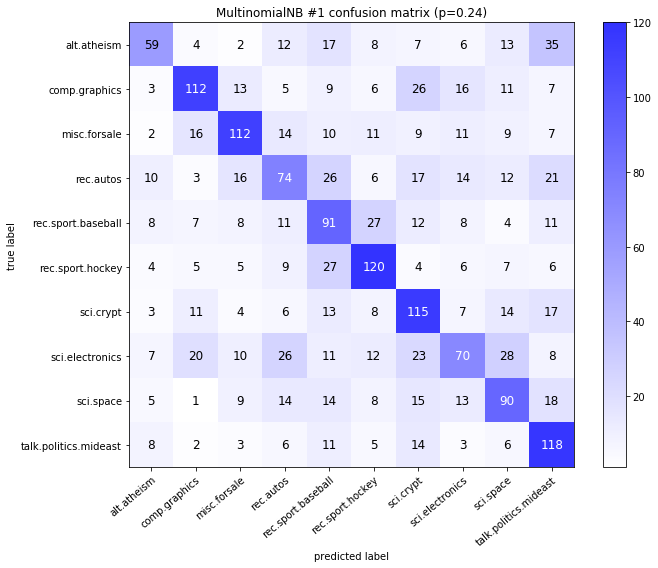
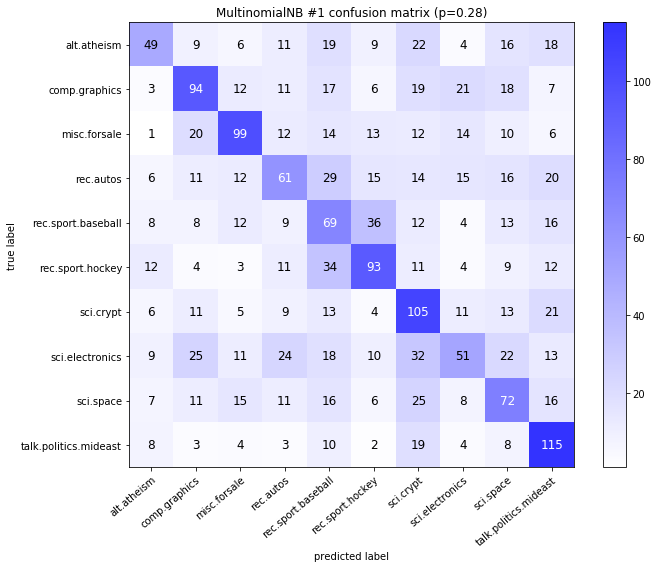
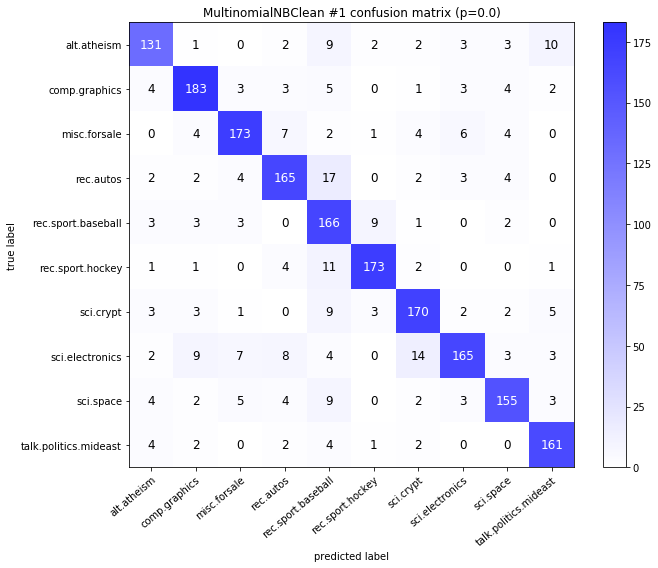
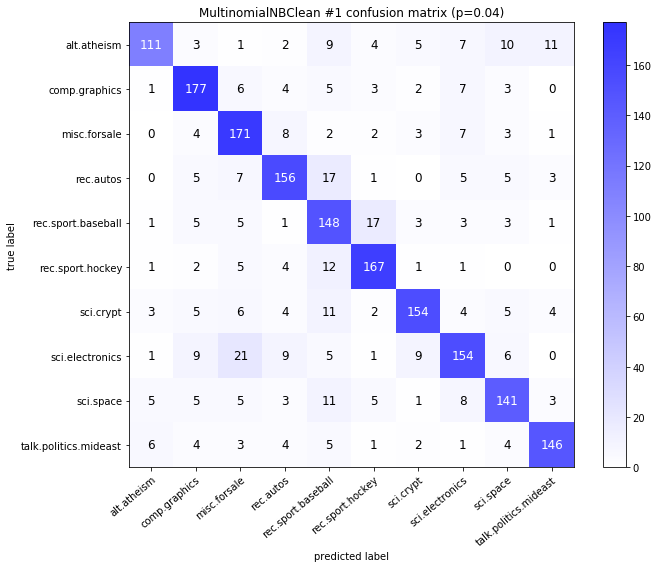
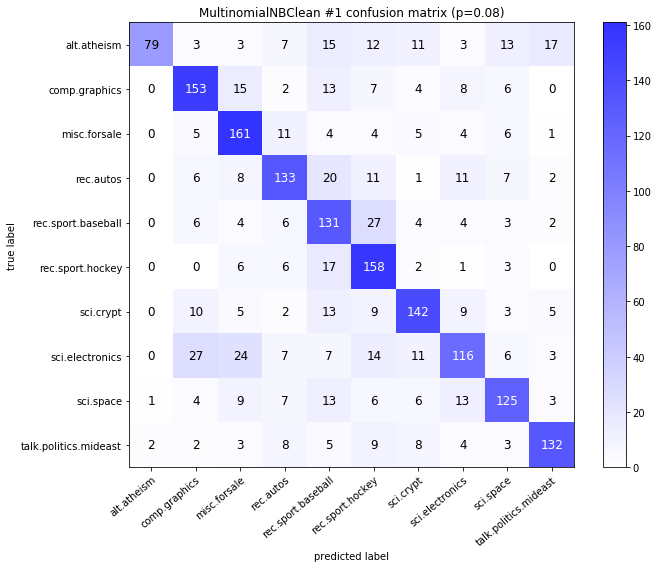
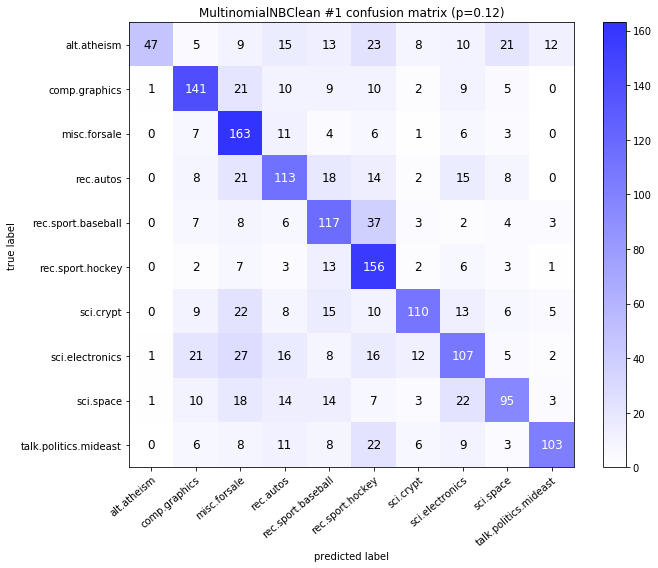
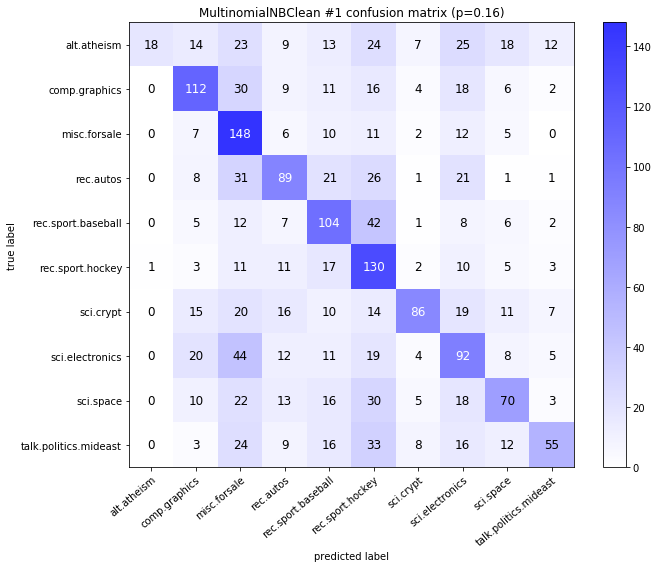
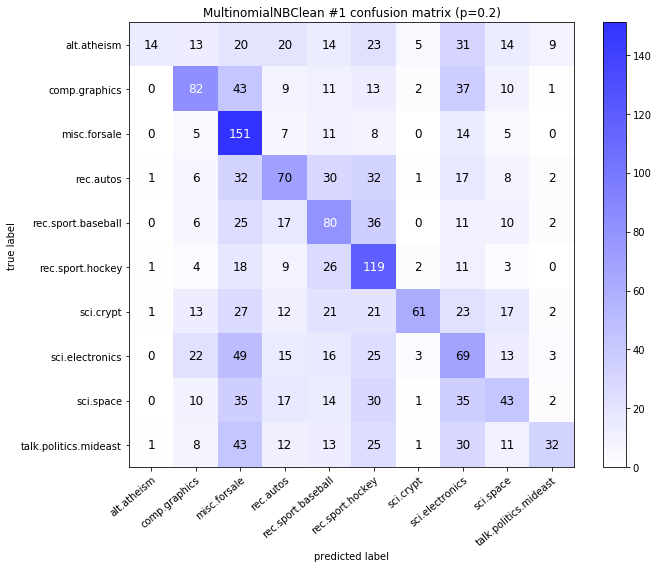
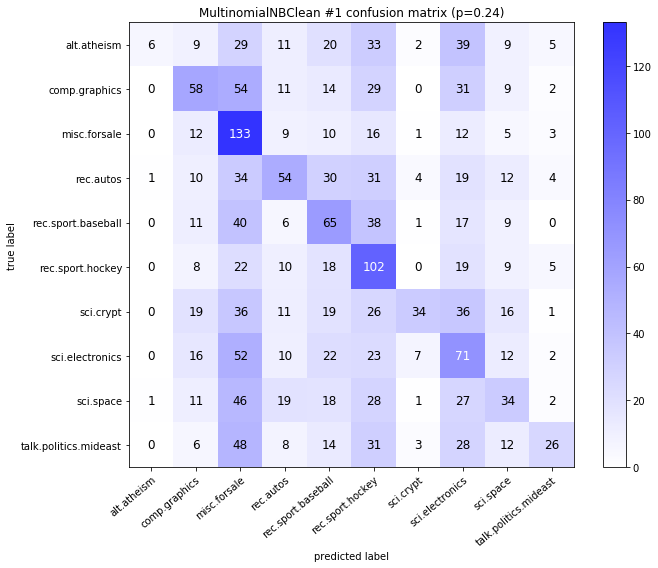
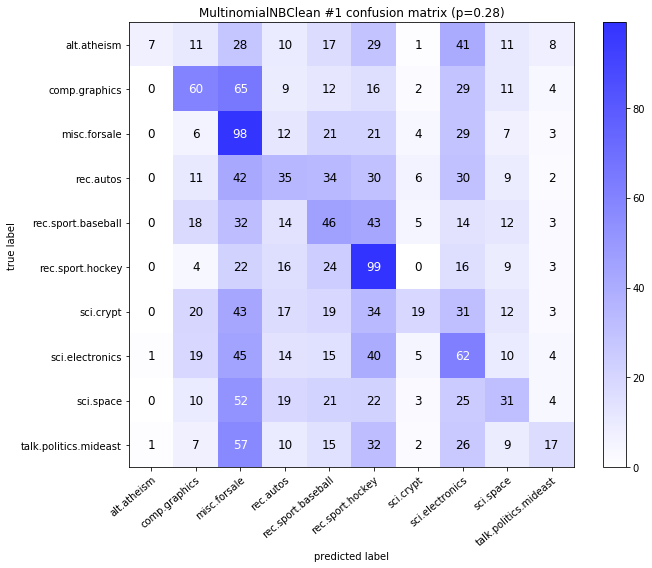
visualize_confusion_matrices(
df,
label_names,
score_name="test_mean_accuracy",
is_higher_score_better=True,
err_param_name="p",
labels_col="test_labels",
predictions_col="predicted_test_labels",
)
plt.show()
[9]:
def main():
train_data, test_data, train_labels, test_labels, label_names, dataset_name = get_data()
df = runner.run(
train_data=train_data,
test_data=test_data,
preproc=Preprocessor,
preproc_params=None,
err_root_node=get_err_root_node(),
err_params_list=get_err_params_list(),
model_params_dict_list=get_model_params_dict_list(train_labels, test_labels),
)
print_results_by_model(df, dropped_columns=[
"train_labels", "test_labels", "reduced_test_data", "confusion_matrix", "predicted_test_labels",
"radius_generator", "missing_value"
])
visualize(df, dataset_name, label_names, test_data)
Models LinearSVCClean and MultinomialNBClean have been trained with clean data and LinearSVC and MultinomialNB with erroneus data.
[10]:
main()
100%|██████████| 8/8 [02:28<00:00, 17.53s/it]
LinearSVC #1
| test_mean_accuracy | train_mean_accuracy | p | C | time_err | time_pre | time_mod | |
|---|---|---|---|---|---|---|---|
| 0 | 0.720 | 0.792343 | 0.00 | 0.001 | 2.937 | 19.858 | 0.198 |
| 1 | 0.809 | 0.884620 | 0.00 | 0.010 | 2.937 | 19.858 | 0.250 |
| 2 | 0.842 | 0.946658 | 0.00 | 0.100 | 2.937 | 19.858 | 0.292 |
| 3 | 0.846 | 0.973005 | 0.00 | 1.000 | 2.937 | 19.858 | 0.468 |
| 4 | 0.835 | 0.974043 | 0.00 | 10.000 | 2.937 | 19.858 | 2.291 |
| 5 | 0.633 | 0.749513 | 0.04 | 0.001 | 27.034 | 20.458 | 0.244 |
| 6 | 0.734 | 0.875016 | 0.04 | 0.010 | 27.034 | 20.458 | 0.282 |
| 7 | 0.780 | 0.948215 | 0.04 | 0.100 | 27.034 | 20.458 | 0.347 |
| 8 | 0.787 | 0.973653 | 0.04 | 1.000 | 27.034 | 20.458 | 0.586 |
| 9 | 0.772 | 0.974173 | 0.04 | 10.000 | 27.034 | 20.458 | 2.702 |
| 10 | 0.537 | 0.695652 | 0.08 | 0.001 | 40.465 | 19.995 | 0.345 |
| 11 | 0.649 | 0.842440 | 0.08 | 0.010 | 40.465 | 19.995 | 0.321 |
| 12 | 0.704 | 0.947696 | 0.08 | 0.100 | 40.465 | 19.995 | 0.336 |
| 13 | 0.706 | 0.973783 | 0.08 | 1.000 | 40.465 | 19.995 | 0.589 |
| 14 | 0.686 | 0.974822 | 0.08 | 10.000 | 40.465 | 19.995 | 2.649 |
| 15 | 0.472 | 0.642440 | 0.12 | 0.001 | 58.590 | 19.594 | 0.198 |
| 16 | 0.589 | 0.823361 | 0.12 | 0.010 | 58.590 | 19.594 | 0.235 |
| 17 | 0.652 | 0.941856 | 0.12 | 0.100 | 58.590 | 19.594 | 0.298 |
| 18 | 0.652 | 0.973913 | 0.12 | 1.000 | 58.590 | 19.594 | 0.528 |
| 19 | 0.617 | 0.974432 | 0.12 | 10.000 | 58.590 | 19.594 | 2.288 |
| 20 | 0.420 | 0.578326 | 0.16 | 0.001 | 82.425 | 20.489 | 0.190 |
| 21 | 0.524 | 0.765347 | 0.16 | 0.010 | 82.425 | 20.489 | 0.237 |
| 22 | 0.598 | 0.924335 | 0.16 | 0.100 | 82.425 | 20.489 | 0.302 |
| 23 | 0.601 | 0.972745 | 0.16 | 1.000 | 82.425 | 20.489 | 0.579 |
| 24 | 0.558 | 0.974043 | 0.16 | 10.000 | 82.425 | 20.489 | 2.857 |
| 25 | 0.352 | 0.537573 | 0.20 | 0.001 | 93.475 | 21.156 | 0.148 |
| 26 | 0.449 | 0.724854 | 0.20 | 0.010 | 93.475 | 21.156 | 0.188 |
| 27 | 0.520 | 0.903310 | 0.20 | 0.100 | 93.475 | 21.156 | 0.237 |
| 28 | 0.516 | 0.968592 | 0.20 | 1.000 | 93.475 | 21.156 | 0.473 |
| 29 | 0.475 | 0.972745 | 0.20 | 10.000 | 93.475 | 21.156 | 2.220 |
| 30 | 0.310 | 0.489552 | 0.24 | 0.001 | 94.942 | 20.544 | 0.131 |
| 31 | 0.376 | 0.655029 | 0.24 | 0.010 | 94.942 | 20.544 | 0.171 |
| 32 | 0.461 | 0.872550 | 0.24 | 0.100 | 94.942 | 20.544 | 0.208 |
| 33 | 0.473 | 0.962362 | 0.24 | 1.000 | 94.942 | 20.544 | 0.459 |
| 34 | 0.420 | 0.971188 | 0.24 | 10.000 | 94.942 | 20.544 | 2.434 |
| 35 | 0.275 | 0.442959 | 0.28 | 0.001 | 125.194 | 17.138 | 0.115 |
| 36 | 0.349 | 0.600130 | 0.28 | 0.010 | 125.194 | 17.138 | 0.147 |
| 37 | 0.420 | 0.838676 | 0.28 | 0.100 | 125.194 | 17.138 | 0.186 |
| 38 | 0.393 | 0.950162 | 0.28 | 1.000 | 125.194 | 17.138 | 0.425 |
| 39 | 0.361 | 0.967683 | 0.28 | 10.000 | 125.194 | 17.138 | 2.252 |
LinearSVCClean #1
| test_mean_accuracy | train_mean_accuracy | p | C | time_err | time_pre | time_mod | |
|---|---|---|---|---|---|---|---|
| 0 | 0.720 | 0.792343 | 0.00 | 0.001 | 2.937 | 19.858 | 0.199 |
| 1 | 0.809 | 0.884620 | 0.00 | 0.010 | 2.937 | 19.858 | 0.253 |
| 2 | 0.842 | 0.946658 | 0.00 | 0.100 | 2.937 | 19.858 | 0.289 |
| 3 | 0.846 | 0.973005 | 0.00 | 1.000 | 2.937 | 19.858 | 0.462 |
| 4 | 0.835 | 0.974043 | 0.00 | 10.000 | 2.937 | 19.858 | 2.554 |
| 5 | 0.658 | 0.792343 | 0.04 | 0.001 | 27.034 | 20.458 | 0.198 |
| 6 | 0.750 | 0.884620 | 0.04 | 0.010 | 27.034 | 20.458 | 0.251 |
| 7 | 0.778 | 0.946658 | 0.04 | 0.100 | 27.034 | 20.458 | 0.290 |
| 8 | 0.771 | 0.973005 | 0.04 | 1.000 | 27.034 | 20.458 | 0.463 |
| 9 | 0.745 | 0.974043 | 0.04 | 10.000 | 27.034 | 20.458 | 2.313 |
| 10 | 0.567 | 0.792343 | 0.08 | 0.001 | 40.465 | 19.995 | 0.230 |
| 11 | 0.677 | 0.884620 | 0.08 | 0.010 | 40.465 | 19.995 | 0.242 |
| 12 | 0.706 | 0.946658 | 0.08 | 0.100 | 40.465 | 19.995 | 0.278 |
| 13 | 0.664 | 0.973005 | 0.08 | 1.000 | 40.465 | 19.995 | 0.443 |
| 14 | 0.625 | 0.974043 | 0.08 | 10.000 | 40.465 | 19.995 | 2.165 |
| 15 | 0.475 | 0.792343 | 0.12 | 0.001 | 58.590 | 19.594 | 0.182 |
| 16 | 0.583 | 0.884620 | 0.12 | 0.010 | 58.590 | 19.594 | 0.232 |
| 17 | 0.608 | 0.946658 | 0.12 | 0.100 | 58.590 | 19.594 | 0.269 |
| 18 | 0.555 | 0.973005 | 0.12 | 1.000 | 58.590 | 19.594 | 0.427 |
| 19 | 0.519 | 0.974043 | 0.12 | 10.000 | 58.590 | 19.594 | 2.482 |
| 20 | 0.396 | 0.792343 | 0.16 | 0.001 | 82.425 | 20.489 | 0.203 |
| 21 | 0.502 | 0.884620 | 0.16 | 0.010 | 82.425 | 20.489 | 0.259 |
| 22 | 0.509 | 0.946658 | 0.16 | 0.100 | 82.425 | 20.489 | 0.298 |
| 23 | 0.451 | 0.973005 | 0.16 | 1.000 | 82.425 | 20.489 | 0.478 |
| 24 | 0.413 | 0.974043 | 0.16 | 10.000 | 82.425 | 20.489 | 3.299 |
| 25 | 0.321 | 0.792343 | 0.20 | 0.001 | 93.475 | 21.156 | 0.181 |
| 26 | 0.409 | 0.884620 | 0.20 | 0.010 | 93.475 | 21.156 | 0.229 |
| 27 | 0.411 | 0.946658 | 0.20 | 0.100 | 93.475 | 21.156 | 0.265 |
| 28 | 0.349 | 0.973005 | 0.20 | 1.000 | 93.475 | 21.156 | 0.425 |
| 29 | 0.312 | 0.974043 | 0.20 | 10.000 | 93.475 | 21.156 | 2.086 |
| 30 | 0.263 | 0.792343 | 0.24 | 0.001 | 94.942 | 20.544 | 0.189 |
| 31 | 0.344 | 0.884620 | 0.24 | 0.010 | 94.942 | 20.544 | 0.232 |
| 32 | 0.348 | 0.946658 | 0.24 | 0.100 | 94.942 | 20.544 | 0.268 |
| 33 | 0.297 | 0.973005 | 0.24 | 1.000 | 94.942 | 20.544 | 0.430 |
| 34 | 0.269 | 0.974043 | 0.24 | 10.000 | 94.942 | 20.544 | 2.112 |
| 35 | 0.220 | 0.792343 | 0.28 | 0.001 | 125.194 | 17.138 | 0.180 |
| 36 | 0.273 | 0.884620 | 0.28 | 0.010 | 125.194 | 17.138 | 0.230 |
| 37 | 0.280 | 0.946658 | 0.28 | 0.100 | 125.194 | 17.138 | 0.265 |
| 38 | 0.243 | 0.973005 | 0.28 | 1.000 | 125.194 | 17.138 | 0.426 |
| 39 | 0.232 | 0.974043 | 0.28 | 10.000 | 125.194 | 17.138 | 2.078 |
MultinomialNB #1
| test_mean_accuracy | train_mean_accuracy | p | alpha | time_err | time_pre | time_mod | |
|---|---|---|---|---|---|---|---|
| 0 | 0.834 | 0.961583 | 0.00 | 0.0001 | 2.937 | 19.858 | 0.035 |
| 1 | 0.843 | 0.960675 | 0.00 | 0.0010 | 2.937 | 19.858 | 0.032 |
| 2 | 0.851 | 0.958209 | 0.00 | 0.0100 | 2.937 | 19.858 | 0.032 |
| 3 | 0.852 | 0.951979 | 0.00 | 0.1000 | 2.937 | 19.858 | 0.032 |
| 4 | 0.832 | 0.925892 | 0.00 | 1.0000 | 2.937 | 19.858 | 0.032 |
| 5 | 0.741 | 0.965737 | 0.04 | 0.0001 | 27.034 | 20.458 | 0.046 |
| 6 | 0.762 | 0.965217 | 0.04 | 0.0010 | 27.034 | 20.458 | 0.040 |
| 7 | 0.782 | 0.964698 | 0.04 | 0.0100 | 27.034 | 20.458 | 0.039 |
| 8 | 0.796 | 0.959507 | 0.04 | 0.1000 | 27.034 | 20.458 | 0.039 |
| 9 | 0.770 | 0.930305 | 0.04 | 1.0000 | 27.034 | 20.458 | 0.039 |
| 10 | 0.671 | 0.964309 | 0.08 | 0.0001 | 40.465 | 19.995 | 0.083 |
| 11 | 0.698 | 0.963920 | 0.08 | 0.0010 | 40.465 | 19.995 | 0.078 |
| 12 | 0.730 | 0.963790 | 0.08 | 0.0100 | 40.465 | 19.995 | 0.079 |
| 13 | 0.750 | 0.960545 | 0.08 | 0.1000 | 40.465 | 19.995 | 0.078 |
| 14 | 0.687 | 0.921739 | 0.08 | 1.0000 | 40.465 | 19.995 | 0.056 |
| 15 | 0.631 | 0.961324 | 0.12 | 0.0001 | 58.590 | 19.594 | 0.036 |
| 16 | 0.659 | 0.961454 | 0.12 | 0.0010 | 58.590 | 19.594 | 0.029 |
| 17 | 0.691 | 0.961064 | 0.12 | 0.0100 | 58.590 | 19.594 | 0.029 |
| 18 | 0.705 | 0.957820 | 0.12 | 0.1000 | 58.590 | 19.594 | 0.029 |
| 19 | 0.629 | 0.908371 | 0.12 | 1.0000 | 58.590 | 19.594 | 0.029 |
| 20 | 0.551 | 0.954835 | 0.16 | 0.0001 | 82.425 | 20.489 | 0.031 |
| 21 | 0.584 | 0.954835 | 0.16 | 0.0010 | 82.425 | 20.489 | 0.029 |
| 22 | 0.614 | 0.954445 | 0.16 | 0.0100 | 82.425 | 20.489 | 0.029 |
| 23 | 0.631 | 0.952888 | 0.16 | 0.1000 | 82.425 | 20.489 | 0.029 |
| 24 | 0.562 | 0.882154 | 0.16 | 1.0000 | 82.425 | 20.489 | 0.029 |
| 25 | 0.475 | 0.944452 | 0.20 | 0.0001 | 93.475 | 21.156 | 0.025 |
| 26 | 0.500 | 0.944322 | 0.20 | 0.0010 | 93.475 | 21.156 | 0.023 |
| 27 | 0.532 | 0.943933 | 0.20 | 0.0100 | 93.475 | 21.156 | 0.023 |
| 28 | 0.553 | 0.939779 | 0.20 | 0.1000 | 93.475 | 21.156 | 0.023 |
| 29 | 0.475 | 0.852304 | 0.20 | 1.0000 | 93.475 | 21.156 | 0.023 |
| 30 | 0.434 | 0.930435 | 0.24 | 0.0001 | 94.942 | 20.544 | 0.026 |
| 31 | 0.441 | 0.930565 | 0.24 | 0.0010 | 94.942 | 20.544 | 0.020 |
| 32 | 0.479 | 0.930954 | 0.24 | 0.0100 | 94.942 | 20.544 | 0.020 |
| 33 | 0.499 | 0.927709 | 0.24 | 0.1000 | 94.942 | 20.544 | 0.020 |
| 34 | 0.419 | 0.814666 | 0.24 | 1.0000 | 94.942 | 20.544 | 0.020 |
| 35 | 0.365 | 0.910578 | 0.28 | 0.0001 | 125.194 | 17.138 | 0.018 |
| 36 | 0.385 | 0.910578 | 0.28 | 0.0010 | 125.194 | 17.138 | 0.017 |
| 37 | 0.404 | 0.909799 | 0.28 | 0.0100 | 125.194 | 17.138 | 0.017 |
| 38 | 0.419 | 0.905775 | 0.28 | 0.1000 | 125.194 | 17.138 | 0.017 |
| 39 | 0.367 | 0.771317 | 0.28 | 1.0000 | 125.194 | 17.138 | 0.017 |
MultinomialNBClean #1
| test_mean_accuracy | train_mean_accuracy | p | alpha | time_err | time_pre | time_mod | |
|---|---|---|---|---|---|---|---|
| 0 | 0.834 | 0.961583 | 0.00 | 0.0001 | 2.937 | 19.858 | 0.035 |
| 1 | 0.843 | 0.960675 | 0.00 | 0.0010 | 2.937 | 19.858 | 0.033 |
| 2 | 0.851 | 0.958209 | 0.00 | 0.0100 | 2.937 | 19.858 | 0.033 |
| 3 | 0.852 | 0.951979 | 0.00 | 0.1000 | 2.937 | 19.858 | 0.032 |
| 4 | 0.832 | 0.925892 | 0.00 | 1.0000 | 2.937 | 19.858 | 0.032 |
| 5 | 0.707 | 0.961583 | 0.04 | 0.0001 | 27.034 | 20.458 | 0.035 |
| 6 | 0.732 | 0.960675 | 0.04 | 0.0010 | 27.034 | 20.458 | 0.033 |
| 7 | 0.769 | 0.958209 | 0.04 | 0.0100 | 27.034 | 20.458 | 0.033 |
| 8 | 0.791 | 0.951979 | 0.04 | 0.1000 | 27.034 | 20.458 | 0.032 |
| 9 | 0.782 | 0.925892 | 0.04 | 1.0000 | 27.034 | 20.458 | 0.033 |
| 10 | 0.520 | 0.961583 | 0.08 | 0.0001 | 40.465 | 19.995 | 0.033 |
| 11 | 0.559 | 0.960675 | 0.08 | 0.0010 | 40.465 | 19.995 | 0.038 |
| 12 | 0.596 | 0.958209 | 0.08 | 0.0100 | 40.465 | 19.995 | 0.045 |
| 13 | 0.665 | 0.951979 | 0.08 | 0.1000 | 40.465 | 19.995 | 0.053 |
| 14 | 0.690 | 0.925892 | 0.08 | 1.0000 | 40.465 | 19.995 | 0.062 |
| 15 | 0.393 | 0.961583 | 0.12 | 0.0001 | 58.590 | 19.594 | 0.030 |
| 16 | 0.417 | 0.960675 | 0.12 | 0.0010 | 58.590 | 19.594 | 0.028 |
| 17 | 0.455 | 0.958209 | 0.12 | 0.0100 | 58.590 | 19.594 | 0.027 |
| 18 | 0.522 | 0.951979 | 0.12 | 0.1000 | 58.590 | 19.594 | 0.028 |
| 19 | 0.598 | 0.925892 | 0.12 | 1.0000 | 58.590 | 19.594 | 0.028 |
| 20 | 0.295 | 0.961583 | 0.16 | 0.0001 | 82.425 | 20.489 | 0.034 |
| 21 | 0.304 | 0.960675 | 0.16 | 0.0010 | 82.425 | 20.489 | 0.031 |
| 22 | 0.326 | 0.958209 | 0.16 | 0.0100 | 82.425 | 20.489 | 0.031 |
| 23 | 0.382 | 0.951979 | 0.16 | 0.1000 | 82.425 | 20.489 | 0.031 |
| 24 | 0.469 | 0.925892 | 0.16 | 1.0000 | 82.425 | 20.489 | 0.031 |
| 25 | 0.235 | 0.961583 | 0.20 | 0.0001 | 93.475 | 21.156 | 0.030 |
| 26 | 0.242 | 0.960675 | 0.20 | 0.0010 | 93.475 | 21.156 | 0.028 |
| 27 | 0.246 | 0.958209 | 0.20 | 0.0100 | 93.475 | 21.156 | 0.028 |
| 28 | 0.293 | 0.951979 | 0.20 | 0.1000 | 93.475 | 21.156 | 0.028 |
| 29 | 0.374 | 0.925892 | 0.20 | 1.0000 | 93.475 | 21.156 | 0.028 |
| 30 | 0.189 | 0.961583 | 0.24 | 0.0001 | 94.942 | 20.544 | 0.031 |
| 31 | 0.195 | 0.960675 | 0.24 | 0.0010 | 94.942 | 20.544 | 0.028 |
| 32 | 0.214 | 0.958209 | 0.24 | 0.0100 | 94.942 | 20.544 | 0.028 |
| 33 | 0.242 | 0.951979 | 0.24 | 0.1000 | 94.942 | 20.544 | 0.028 |
| 34 | 0.303 | 0.925892 | 0.24 | 1.0000 | 94.942 | 20.544 | 0.028 |
| 35 | 0.174 | 0.961583 | 0.28 | 0.0001 | 125.194 | 17.138 | 0.031 |
| 36 | 0.175 | 0.960675 | 0.28 | 0.0010 | 125.194 | 17.138 | 0.028 |
| 37 | 0.182 | 0.958209 | 0.28 | 0.0100 | 125.194 | 17.138 | 0.028 |
| 38 | 0.211 | 0.951979 | 0.28 | 0.1000 | 125.194 | 17.138 | 0.028 |
| 39 | 0.246 | 0.925892 | 0.28 | 1.0000 | 125.194 | 17.138 | 0.028 |
/wrk/users/thalvari/dpEmu/dpemu/plotting_utils.py:299: RuntimeWarning: More than 20 figures have been opened. Figures created through the pyplot interface (`matplotlib.pyplot.figure`) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam `figure.max_open_warning`).
fig, ax = plt.subplots(figsize=(10, 8))

The notebook for this case study can be found here.